New Raman Microscopy Technique Accelerates Digital Pathology
Current methods for pathological diagnosis of diseases require laborious and costly sample processing steps and provide inherently qualitative information. Moreover, batch to batch variations in staining intensity results in highly variable images, making it difficult to use them with automated assessment tools. Marc Kirschner’s lab has developed a novel Raman microscopy method named Normalized Raman Imaging (NoRI), that provides rapid, quantitative, depth-resolved and high-resolution measurements of chemical composition from live or fixed thick tissue samples without the need for any staining. With this method it is possible to generate histology-like images of protein, lipid and water content for digital, remote and quantitative pathology applications, as well as for the study of a wide range of biological mechanisms and diseases.
Advantages
- Absolute concentration measurement of protein, lipid and water.
- Applicable for live or fixed thick tissue samples without the need for staining or sample processing.
- Very high spatial resolution and measurement sensitivity.
- Inherently digital image data suitable for quantitative and remote pathology.
Applications
The unique advantages of NoRI enable a wide range of quantitative pathology applications where it is crucial to obtain quantitative information that can be directly compared between different samples. The information obtained is inherently digital and can be easily transferred and shared for remote pathology applications. Automated assessment tools can be readily developed based on its quantitative information, mitigating numerous limitations associated with distortions and variations of conventional histology images. This platform can also be used for the study of diverse research topics where the amount and distribution of protein and lipid mass is central, including and not limited to cell growth, metabolic diseases, and neurodegenerative diseases.
Current methods for pathological diagnosis of diseases require laborious and costly sample processing steps and provide inherently qualitative information. Moreover, batch to batch variations in staining intensity results in highly variable images, making it difficult to use them with automated assessment tools. Marc Kirschner’s lab has developed a novel Raman microscopy method named Normalized Raman Imaging (NoRI), that provides rapid, quantitative, depth-resolved and high-resolution measurements of chemical composition from live or fixed thick tissue samples without the need for any staining. With this method it is possible to generate histology-like images of protein, lipid and water content for digital, remote and quantitative pathology applications, as well as for the study of a wide range of biological mechanisms and diseases.
Advantages
- Absolute concentration measurement of protein, lipid and water.
- Applicable for live or fixed thick tissue samples without the need for staining or sample processing.
- Very high spatial resolution and measurement sensitivity.
- Inherently digital image data suitable for quantitative and remote pathology.
The unique advantages of NoRI enable a wide range of quantitative pathology applications where it is crucial to obtain quantitative information that can be directly compared between different samples. The information obtained is inherently digital and can be easily transferred and shared for remote pathology applications. Automated assessment tools can be readily developed based on its quantitative information, mitigating numerous limitations associated with distortions and variations of conventional histology images. This platform can also be used for the study of diverse research topics where the amount and distribution of protein and lipid mass is central, including and not limited to cell growth, metabolic diseases, and neurodegenerative diseases.
Intellectual Property Status: Patent(s) Pending
Case Number: 7797
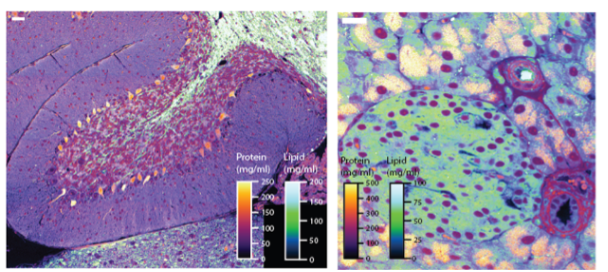
NoRI visualization and measurements of mouse cerebellum and pancreatic islet enabling quantitative pathology. (Credit: Kirschner Lab.)

